

In hopes of generating a simple, concise rule for predicting coreceptor usage, we ranked each of the attributes according to Quinlan's information gain ratio. We built classifiers that employed only the best two sequence positions according to this measure, positions 8 and 12 in our alignment (7 and 11 in the public consensus).
Below is a condensed version of a decision tree produced using only these two positions. Based on cross-validation experiments, this tree is expected to classify unseen test cases with 89.97% accuracy. It classifies 246 out of our 271 training cases correctly (90.77%). Here is a breakdown of its performance in cross-validation.| TP Rate | FP Rate | Precision | Recall | Class |
| 0.757 | 0.012 | 0.975 | 0.757 | cxcr4 |
| 0.988 | 0.243 | 0.869 | 0.988 | ccr5 |
Here is a somewhat condensed version of the decision tree produced in WEKA.
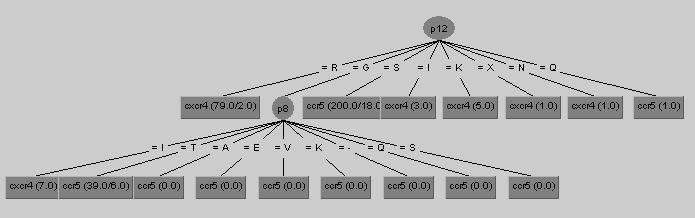
Which at the moment would look like this in the WetCat window:
J48 pruned tree ------------------ p12 = R: cxcr4 (69.0/2.0) p12 = G | p8 = I: cxcr4 (4.0) | p8 = T: ccr5 (28.0/6.0) p12 = S: ccr5 (160.0/17.0) p12 = I: cxcr4 (3.0) p12 = K: cxcr4 (4.0) p12 = X: cxcr4 (1.0) p12 = N: cxcr4 (1.0) p12 = Q: ccr5 (1.0)
If translated into english, both of these representations would read "If there is an R at position 12, then the class for the sequence is CXCR4, otherwise if there is a G at position 12 and a I at position 8 then CXCR4, otherwise if there is a G at position 12 and a T at position 8 then CCR5 ..." The numbers in parantheses to the right of the leaves indicate how many samples from the training set reached that leaf versus how many were incorrectly classified. (# reached / # misclassified) .